Gene Regulatory Networks
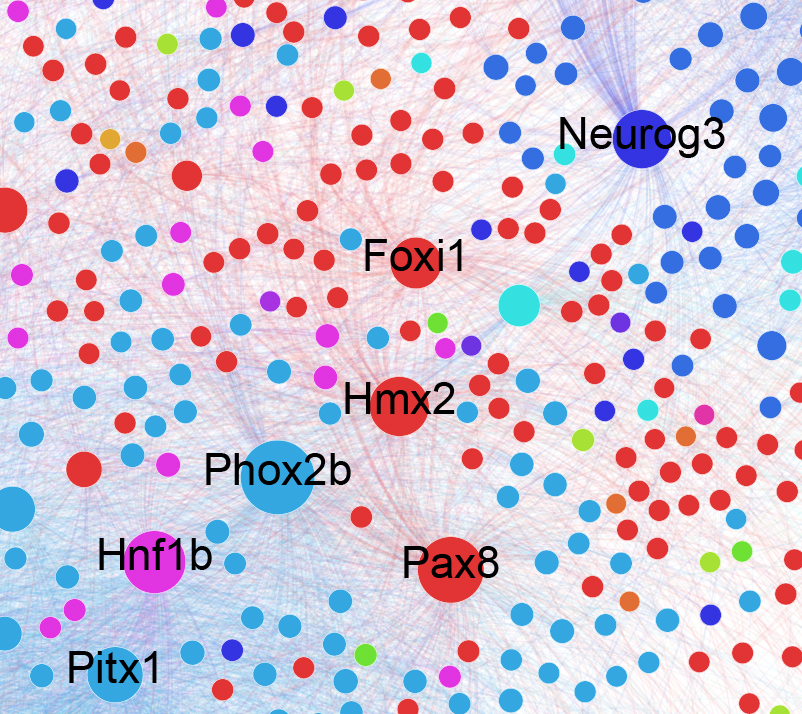
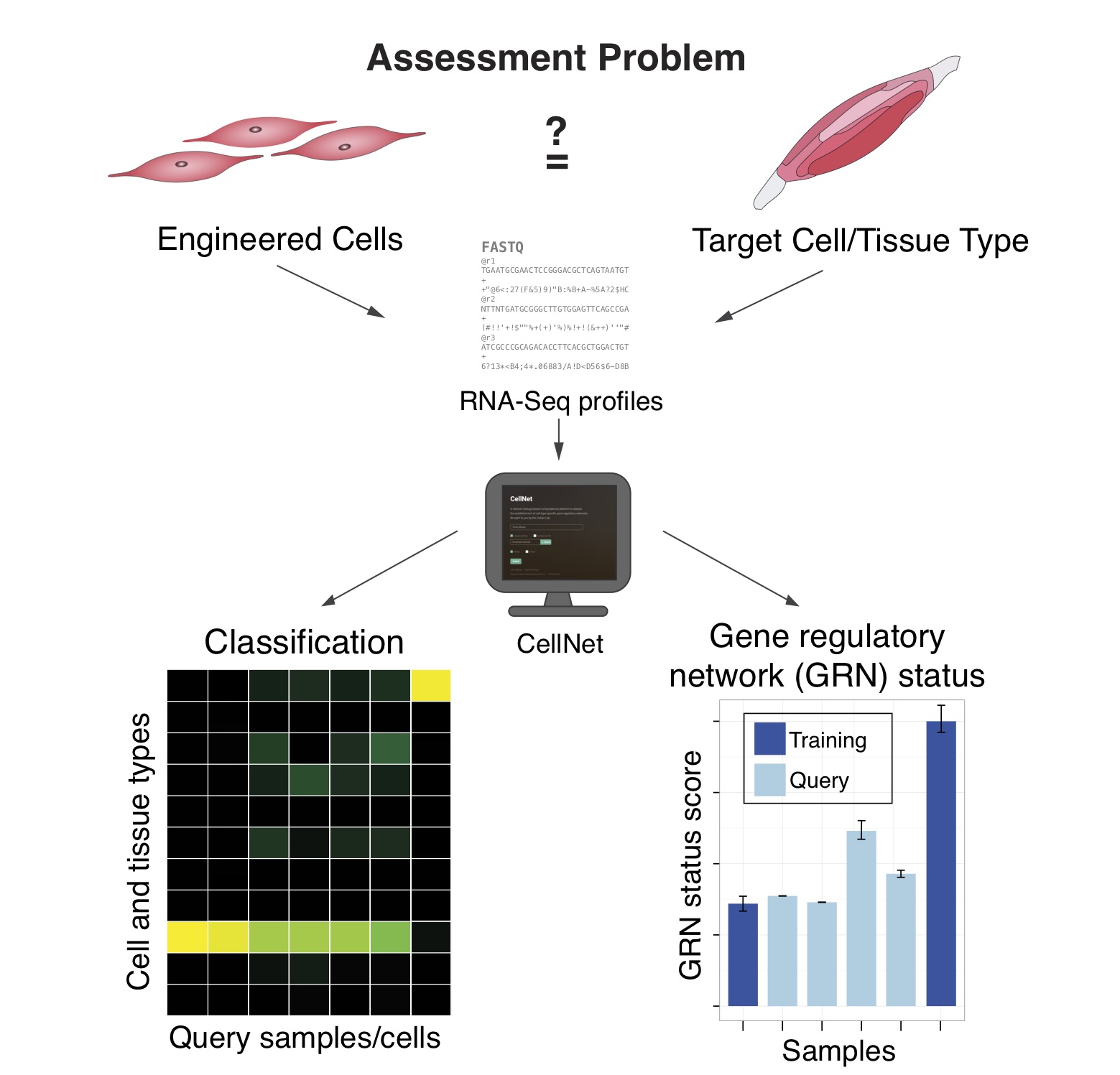
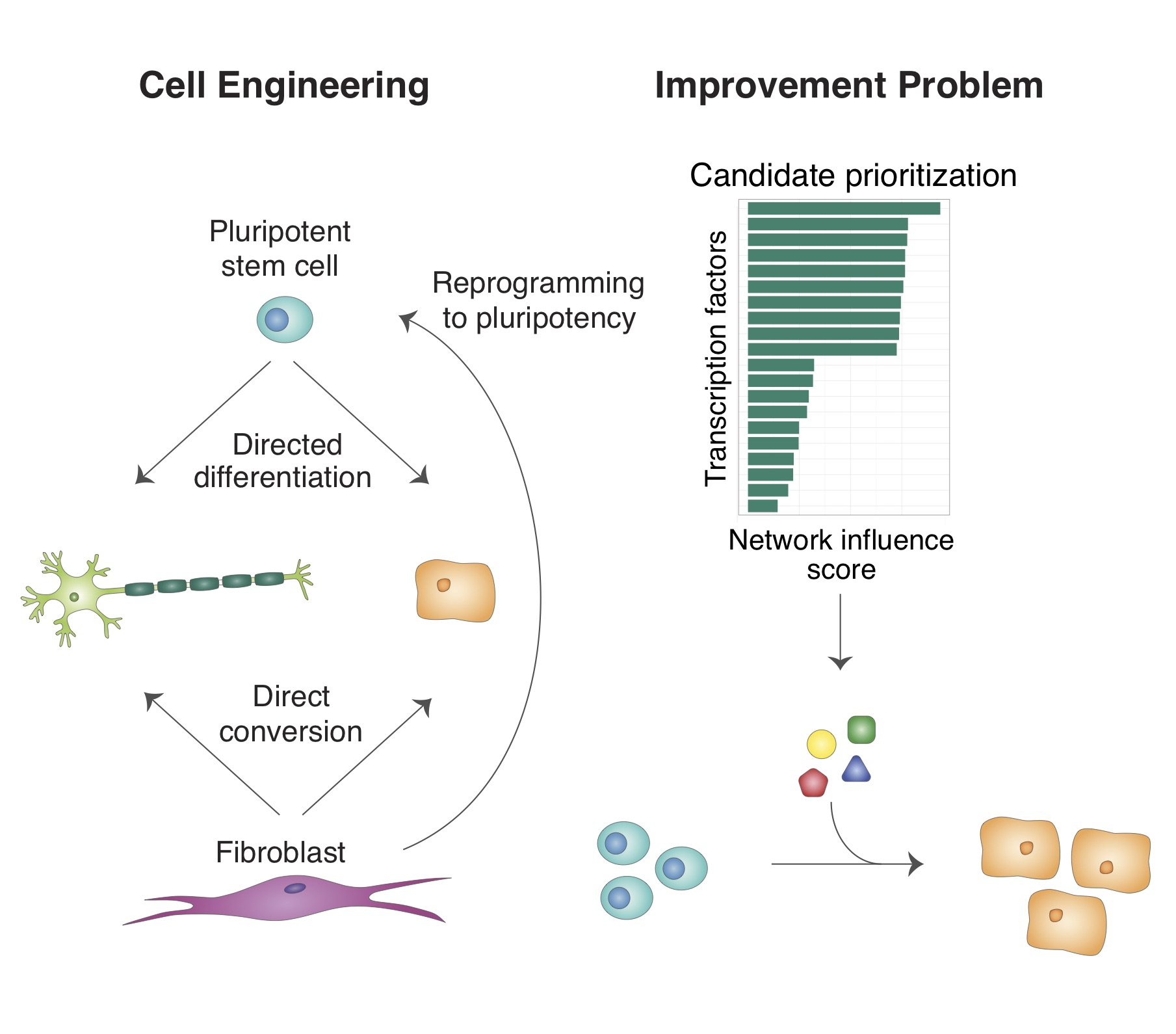
Gene regulatory networks (GRNs), encoded in the genome of an organism, define the complete set of regulatory relationships among genes and gene products. GRNs govern the cell’s transcriptional output both at steady state and in response to perturbations, and thus act as major molecular determinants of cell-type identity. The study of GRNs is a central and unifying component of our research program. We are actively developing new algorithms to reconstruct GRNs, to measure their establishment, to infer their dynamics, and to model intercellular regulatory networks. We are especially interested in understanding how cell type specific GRNs are established during synovial joint development.